| Parameter | Mand | Type | Default | Constraints |
| inputlistsets | yes | filename(s) | inputlistset.fits | |
EPIC emldetect
source list(s).
Specifying more than one list, means these lists will be merged.
|
| outputlistset | yes | filename | summarylist.fits | |
FITS output observation source list
|
| htmloutput | yes | filename | summarylist.html | |
Output observation source list in HTML format
|
| crossidsets | no | filename(s) | crossid.fits | |
Source lists from earlier stages of the detection pipeline.
These files will be used to fill in the pointers to the source identification
numbers in these lists.
|
| usecrossidsets | no | Boolean | no | |
Controls whether a cross correlation with X-ray source list from earlier detection stages should be performed.
|
| omlistset | no | filename | omlist.fits | |
OM observation source list. Setting omlistset=- means that no OM data set is available.
|
| useomlistset | no | Boolean | no | |
Controls whether a cross correlation with an OM source list should be performed.
|
| systerr | no | float | 1.0 | [0.0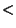 param param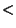 6.0] 6.0] |
Systematic error for EPIC source matching [arcsec]
|
| maxerr | no | float | 1.0 | [3.0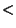 param param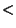 5.0] 5.0] |
Maximum allowed error (stat. + syst.) for EPIC source matching [sigmas].
|
| omdist | no | float | 15.0 | [0.0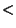 param param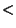 60.0] 60.0] |
Max. distance EPIC-OM for correlation [arcsec]
|
| Parameter | Mand | Type | Default | Constraints |